Chapter 11 Geospatial analysis
This section deals with the use of geospatial analysis in healthcare. There is a detailed tutorial online available here
11.1 Geocoding
There are several packages avalable for obtaining geocode or longitude and latitude of location. The tmaptools package provide free geocoding using OpenStreetMap or OSM overpass API. Both ggmap and googleway access Google Maps API and will require a key and payment for access.
11.1.1 OpenStreetMap
This is a simple example to obtain the geocode of Monash Medical Centre. However, such a simple example does not always work without the full address. There are other libraries for accessing other data from OSM such as parks, restaurants etc.
## $query
## [1] "monash medical centre, clayton"
##
## $coords
## x y
## 145.12357 -37.92001
##
## $bbox
## xmin ymin xmax ymax
## 145.12052 -37.92196 145.12760 -37.91810The osmdata library includes function opg for extracting data from Overpass query. The list is available at https://wiki.openstreetmap.org/wiki/Map_features#Transportation
## -- Attaching packages --------------------------------------- tidyverse 1.3.0 --## v ggplot2 3.3.5 v purrr 0.3.4
## v tibble 3.1.6 v dplyr 1.0.8
## v tidyr 1.1.4 v stringr 1.4.0
## v readr 1.4.0 v forcats 0.5.0## Warning: package 'ggplot2' was built under R version 4.0.5## Warning: package 'tibble' was built under R version 4.0.5## Warning: package 'tidyr' was built under R version 4.0.5## Warning: package 'dplyr' was built under R version 4.0.5## -- Conflicts ------------------------------------------ tidyverse_conflicts() --
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()## Warning: package 'osmdata' was built under R version 4.0.5## Data (c) OpenStreetMap contributors, ODbL 1.0. https://www.openstreetmap.org/copyright## Linking to GEOS 3.8.0, GDAL 3.0.4, PROJ 6.3.111.1.2 Google Maps API
The equivalent code in ggmap is provided below. Note that a key is required from Google Maps API.
library(ggmap)
register_google(key="Your Key")
#geocode
geocode ("monash medical centre, clayton")
#trip
#mapdist("5 stud rd dandenong","monash medical centre")The next demonstration is the extraction of geocodes from multiple addresses embedded in a column of data within a data frame. This is more efficient compared to performing geocoding line by line. An example is provided on how to create your own icon on the leaflet document as well as taking a picture for publication.
library(dplyr)
library(tidyr) #unite verb from tidyr
library(readr)
library(tmaptools)
library(leaflet)
library(sf)
clinic<-read_csv("./Data-Use/TIA_clinics.csv")##
## -- Column specification --------------------------------------------------------
## cols(
## City = col_character(),
## Country = col_character(),
## Setting = col_character(),
## `Pre-COVID-Assessment` = col_character(),
## `Post-COVID-Assessment` = col_character(),
## `Pre-COVID-Imaging` = col_character(),
## `Post-COVID-Imaging` = col_character(),
## `PPE use in clinic` = col_character(),
## `COVID Alert Level` = col_double(),
## `Clinic-Status` = col_character(),
## Date = col_character()
## )clinic2<-clinic %>%
unite ("address",City:Country,sep = ",")%>% filter(!is.na(`Clinic-Status`))
load("./Data-Use/TIAclinics_geo.Rda")
clinics_geo<-left_join(clinics_geo,clinic2, by=c("query"="address"))
#create icon markers
#icon markers
icons_blue <- awesomeIcons(
icon= 'medkit',
iconColor = 'black',
library = 'ion',
markerColor = "blue"
)
icons_red <- awesomeIcons(
icon= 'medkit',
iconColor = 'black',
library = 'ion',
markerColor = "red"
)
#subset
clinics_geo_active<-clinics_geo %>%filter(`Clinic-Status`=="Active")
clinics_geo_inactive<-clinics_geo %>%filter(`Clinic-Status` !="Active")
m<-leaflet(data=clinics_geo) %>%
addTiles() %>% #default is OSM
addAwesomeMarkers(lat=clinics_geo_active$lat,lng=clinics_geo_active$lon,
icon=icons_blue,label = ~as.character(clinics_geo_active$query) ) %>%
addAwesomeMarkers(lat=clinics_geo_inactive$lat,lng=clinics_geo_inactive$lon,
icon=icons_red,label = ~as.character(clinics_geo_inactive$query))
#make pics using mapshot
mapview::mapshot(m, url = paste0(getwd(),
file="./Data-Use/TIAclinic_world.html"), file = paste0(getwd(), "./Data-Use/TIAclinic_world.png"))
mGoogleway has the flexibility of easily interrogating Google Maps API for time of trip and traffic condition.
11.2 Sp and sf objects
There are several methods for reading the shapefile data. Previously rgdal library was used. This approach creates files which can be described as S4 object in that there are slots for different data. The spatial feature sf approach is much easier to handle and the data can easily be subset and merged if needed. An example of conversion between sp and sf is provided.
Here, base R plot is used to illustrate the shapefile of Melbourne and after parcellation into Voronois, centred by the hospital location.
## Loading required package: raster## Loading required package: sp##
## Attaching package: 'raster'## The following object is masked from 'package:dplyr':
##
## select## The following object is masked from 'package:tidyr':
##
## extractlibrary(ggvoronoi)
library(tidyverse)
library(sf)
par(mfrow=c(1,2)) #plot 2 objects in 1 row
msclinic<-read.csv("./Data-Use/msclinic.csv") %>% filter(clinic==1, metropolitan==1)
#convert to spatialpointdataframe
coordinates(msclinic) <- c("lon", "lat")
#proj4string(msclinic) <- CRS("+proj=longlat +datum=WGS84")
proj4string(msclinic) <- CRS("+proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs")
#create voronoi from msclinic data
#msclinic object is in sp
msv<-voronoi(msclinic)
#create voronoi from msclinic data
#object is in sp
msv<-voronoi(msclinic)
#subset Greater Melbourne
Melb<-st_read("./Data-Use/GCCSA_2016_AUST.shp") %>% filter(STE_NAME16=="Victoria",GCC_NAME16=="Greater Melbourne")## Reading layer `GCCSA_2016_AUST' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\GCCSA_2016_AUST.shp' using driver `ESRI Shapefile'
## Simple feature collection with 34 features and 5 fields (with 18 geometries empty)
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: 96.81694 ymin: -43.74051 xmax: 167.998 ymax: -9.142176
## geographic CRS: GDA94#transform sf to sp object
Melb<-as(Melb,Class="Spatial")
plot(Melb)
#voronoi bound by greater melbourne
vor <- dismo::voronoi(msclinic, ext=extent(Melb))
#intersect is present in base R, dplyr, raster, lubridate etc
r <- raster::intersect(vor, Melb)
#r<-intersect(msv,gccsa_fsp)
#
msclinic$hospital<-as.character(msclinic$hospital)
plot(r, col=rainbow(length(r)), lwd=3)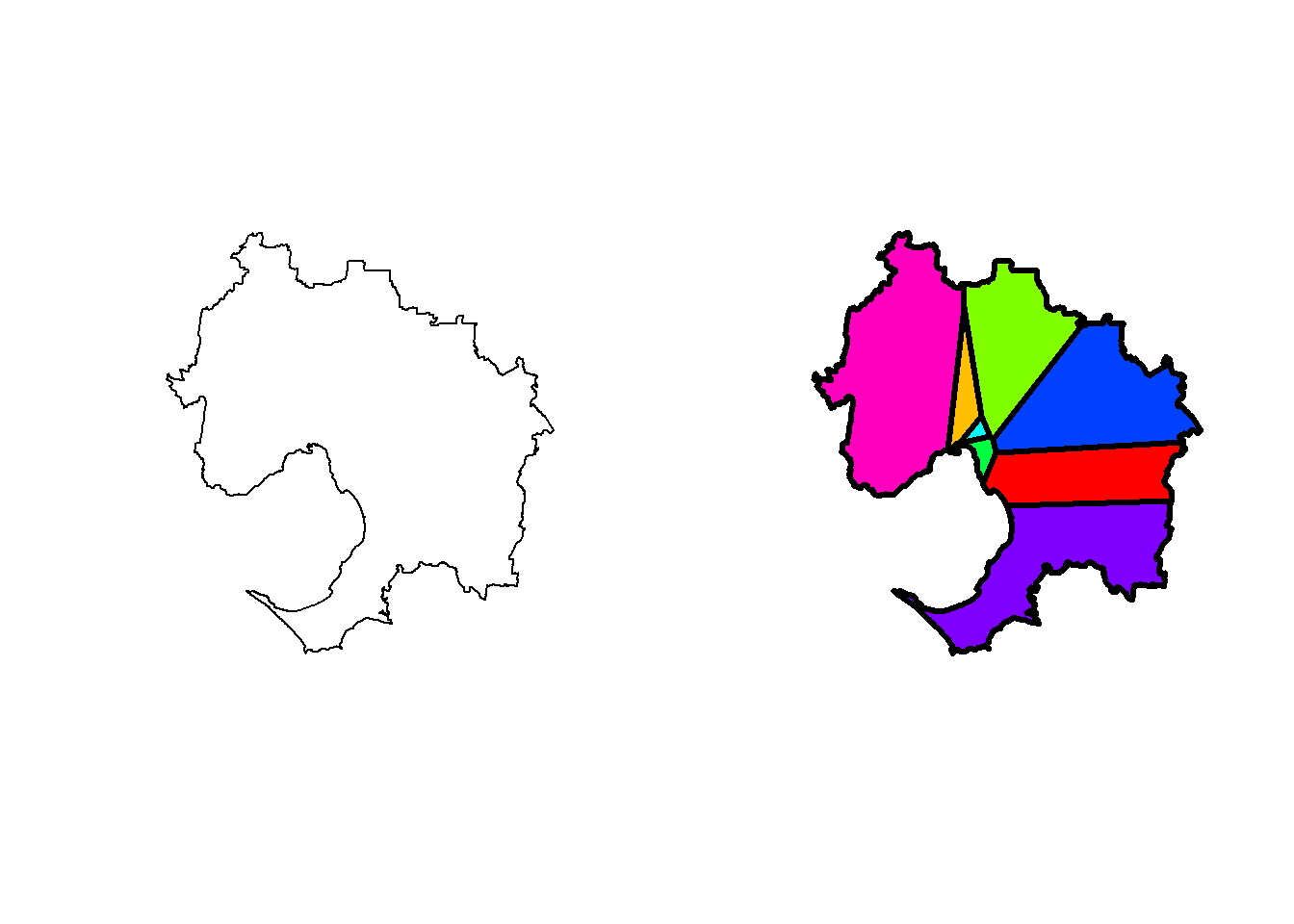
11.2.1
Obtain the Brisbane data as sf object.
## [1] "osm_id" "name" "Size"
## [4] "addr.city" "addr.housenumber" "addr.postcode"
## [7] "addr.state" "addr.street" "addr.suburb"
## [10] "addr.unit" "amenity" "barrier"
## [13] "brand" "building" "capacity"
## [16] "community_centre.for" "contact.phone" "description"
## [19] "email" "entrance" "image"
## [22] "indoor" "level" "name.en"
## [25] "opening_hours" "operator" "operator.type"
## [28] "phone" "source" "source.date"
## [31] "website" "wheelchair" "wheelchair.description"
## [34] "geometry"Extract the centroid and polygons. Check that the coordinate reference system is EPSG 4326 or World Geodetic System (WGS) 84.
ComPoint<-ComCentre$osm_points %>% filter(amenity=="community_centre")
ComPoly <- ComCentre$osm_polygons %>%
st_centroid()## Warning in st_centroid.sf(.): st_centroid assumes attributes are constant over
## geometries of x## Warning in st_centroid.sfc(st_geometry(x), of_largest_polygon =
## of_largest_polygon): st_centroid does not give correct centroids for longitude/
## latitude data## Coordinate Reference System:
## User input: EPSG:4326
## wkt:
## GEOGCRS["WGS 84",
## DATUM["World Geodetic System 1984",
## ELLIPSOID["WGS 84",6378137,298.257223563,
## LENGTHUNIT["metre",1]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433]],
## CS[ellipsoidal,2],
## AXIS["geodetic latitude (Lat)",north,
## ORDER[1],
## ANGLEUNIT["degree",0.0174532925199433]],
## AXIS["geodetic longitude (Lon)",east,
## ORDER[2],
## ANGLEUNIT["degree",0.0174532925199433]],
## USAGE[
## SCOPE["unknown"],
## AREA["World"],
## BBOX[-90,-180,90,180]],
## ID["EPSG",4326]]Plot the map of community centres in Ballarat with tmap. The view argument in tmap_mode set this plotting to interactive viewing with tmap with leaflet. The leaflet library can be called directly using tmap_leaflet
#extract bounding box for Brisbane
Ballarat <- osmdata::getbb("Brisbane, australia", format_out = "sf_polygon")$multipolygon
library(tmap)
#set interactive view
tmap_mode("view")## tmap mode set to interactive viewing11.3 Thematic map
In the first chapter we provided a thematic map example with ggplot2. here we will illustrate with mapview library using open data on Finland.
##
## geofi R package: tools for open GIS data for Finland.
## Part of rOpenGov <ropengov.org>.## Warning: package 'pxweb' was built under R version 4.0.5## pxweb: R tools for PX-WEB API.
## Copyright (C) 2014-2021 Mans Magnusson, Leo Lahti et al.
## https://github.com/ropengov/pxweb## Warning: package 'janitor' was built under R version 4.0.5##
## Attaching package: 'janitor'## The following object is masked from 'package:raster':
##
## crosstab## The following objects are masked from 'package:stats':
##
## chisq.test, fisher.test#data on Finland
#https://pxnet2.stat.fi/PXWeb/pxweb/en/Kuntien_avainluvut/
#Kuntien_avainluvut__2021/kuntien_avainluvut_2021_viimeisin.px/table/
#tableViewLayout1/
FinPop<-readxl::read_xlsx("./Data-Use/Kuntien avainluvut_20210704-153529.xlsx", skip = 2)## New names:
## * `` -> ...1FinPop2<-FinPop[-c(1),] #remove data for entire Finland
#get shapefile for municipality
municipalities <- get_municipalities(year = 2020, scale = 4500) #sf object)## Requesting response from: http://geo.stat.fi/geoserver/wfs?service=WFS&version=1.0.0&request=getFeature&typename=tilastointialueet%3Akunta4500k_2020## Warning: Coercing CRS to epsg:3067 (ETRS89 / TM35FIN)## Data is licensed under: Attribution 4.0 International (CC BY 4.0)#join shapefile and population data
municipalities2<-right_join(municipalities, FinPop2, by=c("name"="...1")) %>%
rename(Pensioner=`Proportion of pensioners of the population, %, 2019`,Age65=`Share of persons aged over 64 of the population, %, 2020`) %>% na.omit()
#plot map using mapview
mapview::mapview(municipalities2["Age65"],layer.name="Percentage Age>65")This example illustrates how to add arguments in mapview
library(mapview)
library(sf)
library(tmaptools)
#NY Shape file
NYsf<-st_read("./Data-Use/Borough_Boundaries/geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0.shp")## Reading layer `geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\Borough_Boundaries\geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0.shp' using driver `ESRI Shapefile'
## Simple feature collection with 5 features and 4 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -74.25559 ymin: 40.49612 xmax: -73.70001 ymax: 40.91553
## geographic CRS: WGS84(DD)#NY subway -does not go to Staten Island
NYsubline<-st_read("./Data-Use/NYsubways/geo_export_147781bc-e472-4c12-8cd2-5f9859f90706.shp")## Reading layer `geo_export_147781bc-e472-4c12-8cd2-5f9859f90706' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\NYsubways\geo_export_147781bc-e472-4c12-8cd2-5f9859f90706.shp' using driver `ESRI Shapefile'
## Simple feature collection with 742 features and 6 fields
## geometry type: LINESTRING
## dimension: XY
## bbox: xmin: -74.03088 ymin: 40.57559 xmax: -73.75541 ymax: 40.90312
## geographic CRS: WGS84(DD)#NY subway stations
NYsubstation<-st_read("./Data-Use/NYsubways/geo_export_0dab2fcf-79b8-409a-b940-7c98778a4418.shp")## Reading layer `geo_export_0dab2fcf-79b8-409a-b940-7c98778a4418' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\NYsubways\geo_export_0dab2fcf-79b8-409a-b940-7c98778a4418.shp' using driver `ESRI Shapefile'
## Simple feature collection with 473 features and 5 fields
## geometry type: POINT
## dimension: XY
## bbox: xmin: -74.03088 ymin: 40.57603 xmax: -73.7554 ymax: 40.90313
## geographic CRS: WGS84(DD)#list of NY hospitals
Hosp<-c("North Shore University Hospital","Long Island Jewish Medical Centre","Staten Island University Hospital","Lennox Hill Hospital","Long Island Jewish Forest Hills","Long Island Jewish Valley Stream","Plainview Hospital","Cohen Children's Medical Center","Glen Cove Hospital","Syosset Hospital")
#geocode NY hospitals and return sf object
Hosp_geo<-geocode_OSM(paste0(Hosp,",","New York",",","USA"),as.sf = TRUE)## No results found for "Long Island Jewish Medical Centre,New York,USA".## No results found for "Lennox Hill Hospital,New York,USA".## No results found for "Cohen Children's Medical Center,New York,USA".#data from jama paper on variation in mortality from covid
mapview(NYsf, zcol="boro_name")+
mapview(NYsubline, zol="name")+
#cex is the circle size default=6
mapview(NYsubstation, zol="line",cex=1)+
mapview(Hosp_geo, zcol="query", cex=3)The example shown under Data wrangling on how to extract data from pdf is now put to use to create thematic map of stroke number per region in Denmark.
library(dplyr)
library(tidyverse)
library(sf)
library(eurostat)
library(leaflet)
library(mapview)
library(tmaptools)
#####################code to generate HospLocations.Rda
load ("./Data-Use/europeRGDK.Rda")
#create data on hospitals
#hosp_addresses <- c(
# AarhusHospital = "aarhus university hospital, aarhus, Denmark",
# AalborgHospital = "Hobrovej 18-22 9100 aalborg, Denmark",
# HolstebroHospital = "Lægårdvej 12 7500 Holstebro, Holstebro, Denmark", VejleHospital="Vejle Sygehus,Beriderbakken 4, Vejle, Denmark",
# EsbjergHospital="Esbjerg Sygehus, Esbjerg, Denmark",
#SoenderborgHospital="Sydvang 1C 6400 Sønderborg, Denmark",
#OdenseHospital="Odense Sygehus, Odense, Denmark",
#RoskildeHospital="Roskilde Sygehus, Roskilde, Denmark",
#BlegdamsvejHospital="Rigshospitalet blegdamsvej, 9 Blegdamsvej, København, Denmark",
#GlostrupHospital="Rigshospitalet Glostrup, Glostrup, Denmark")
#geocode hospitals using OpenStreetMap. This function works better with street addresses
#HospLocations <- tmaptools::geocode_OSM(hosp_addresses, as.sf=TRUE)
#convert data into sf object
#HospLocations <- sf::st_transform(HospLocations, sf::st_crs(europeRGDK))
#CSC comprehensive stroke centre
#PSC primary stroke centre
#HospLocations$Center<-c("CSC", "PSC", "PSC", "PSC", "PSC", "PSC", "CSC", "PSC", "CSC", "PSC")
#save HospLocations
#save(HospLocations, "HospLocations.Rda")
load("./Data-Use/HospLocations.Rda")
##https://ec.europa.eu/eurostat/web/nuts/background
load("./Data-Use/euro_nuts2_sf.Rda")
DKnuts2_sf<- euro_nuts2_sf%>% filter(str_detect(NUTS_ID,"^DK"))
#convert pdf to csv file
dk<-read.csv("./Data-Use/denmarkstrokepdf.csv")
#extract only data on large regions=NUTS2
dk2<-dk[c(4:8),]
#clean up column X.1 containing stroke data
#remove numerator before back slash then remove number before slash sign
dk2$strokenum<-str_replace(dk2$X.1,"[0-9]*","") %>%
str_replace("/","\\") %>% as.numeric()
dk2$Uoplyst<-str_replace(dk2$Uoplyst,"Sjælland","Sjælland")
#merge sf file for DK nuts2 with stroke number
DKnuts2_sf2<-right_join(DKnuts2_sf,dk2,by=c("NUTS_NAME"="Uoplyst"))
#add population from Statistics Denmark 2018
DKnuts2_sf2$pop<-c(589148,1822659,835024, 1313596,1220763)
DKnuts2_sf2$male<-c(297679,894553,416092,657817,610358)
DKnuts2_sf2$maleper<-round(with(DKnuts2_sf2,pop/male),2)
#plot map
mapview(DKnuts2_sf2["strokenum"], layer.name="Stroke Number") +mapview(HospLocations, zcol="Center", layer.name="Hospital Designation")11.3.1 Calculate distance to Hospital-OpenStreetMap
Determine distance hospital to centroid of kommunes rather than the larger regions of Denmark. This can be performed with OpenStreetMap or Google Maps API.
dist_to_loc <- function (geometry, location){
units::set_units(st_distance(st_centroid (geometry), location)[,1], km)
}
#set distance 10 km
#change to 100 km
dist_range <- units::set_units(100, km)
##
europeRGDK <- mutate(europeRGDK,
DirectDistanceToAarhus = dist_to_loc(geometry,HospLocations["AarhusHospital", ]),
DirectDistanceToAalborg = dist_to_loc(geometry,HospLocations["AalborgHospital", ]),
DirectDistanceToHolstebro = dist_to_loc(geometry,HospLocations["HolstebroHospital", ]),
DirectDistanceToVejle = dist_to_loc(geometry,HospLocations["VejleHospital", ]),
DirectDistanceToEsbjerg = dist_to_loc(geometry,HospLocations["EsbjergHospital", ]),
DirectDistanceToSoenderborg = dist_to_loc(geometry,HospLocations["SoenderborgHospital", ]),
DirectDistanceToOdense = dist_to_loc(geometry,HospLocations["OdenseHospital", ]),
DirectDistanceToRoskilde = dist_to_loc(geometry,HospLocations["RoskildeHospital", ]),
DirectDistanceToBlegdamsvej = dist_to_loc(geometry,HospLocations["BlegdamsvejHospital", ]),
DirectDistanceToGlostrup = dist_to_loc(geometry,HospLocations["GlostrupHospital", ]),
#
DirectDistanceToNearest = pmin(DirectDistanceToAarhus,
DirectDistanceToAalborg,DirectDistanceToHolstebro,
DirectDistanceToVejle,DirectDistanceToEsbjerg, DirectDistanceToSoenderborg,DirectDistanceToOdense,
DirectDistanceToRoskilde,DirectDistanceToBlegdamsvej,
DirectDistanceToGlostrup))
StrokeHosp <- filter(europeRGDK, DirectDistanceToNearest < dist_range) %>%
mutate(Postcode = as.numeric(COMM_ID))
head(StrokeHosp)## Simple feature collection with 6 features and 15 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: 8.709358 ymin: 56.83344 xmax: 8.940572 ymax: 56.98483
## CRS: +proj=longlat +ellps=GRS80 +no_defs
## COMM_ID Shape_Leng Shape_Area geometry
## 1 DK10817738668 0.3605216 0.0030530589 MULTIPOLYGON (((8.940572 56...
## 2 DK10817738669 0.1539856 0.0012336619 MULTIPOLYGON (((8.908073 56...
## 3 DK10817738670 0.1630955 0.0009363543 MULTIPOLYGON (((8.886687 56...
## 4 DK10817738671 0.2010890 0.0019239206 MULTIPOLYGON (((8.822215 56...
## 5 DK10817738672 0.1926252 0.0011456026 MULTIPOLYGON (((8.750229 56...
## 6 DK10817738673 0.1793186 0.0011254477 MULTIPOLYGON (((8.83318 56....
## DirectDistanceToAarhus DirectDistanceToAalborg DirectDistanceToHolstebro
## 1 117.9985 [km] 68.34687 [km] 64.51209 [km]
## 2 116.0525 [km] 66.18672 [km] 64.58528 [km]
## 3 114.3120 [km] 67.98462 [km] 60.81974 [km]
## 4 118.4093 [km] 73.88461 [km] 59.18484 [km]
## 5 119.8000 [km] 77.13285 [km] 57.41624 [km]
## 6 114.3753 [km] 72.63258 [km] 56.00828 [km]
## DirectDistanceToVejle DirectDistanceToEsbjerg DirectDistanceToSoenderborg
## 1 141.1704 [km] 163.6407 [km] 231.0773 [km]
## 2 140.0296 [km] 163.5219 [km] 230.0249 [km]
## 3 136.9248 [km] 159.7873 [km] 226.8296 [km]
## 4 138.4835 [km] 158.5284 [km] 228.0678 [km]
## 5 138.2878 [km] 156.8171 [km] 227.6471 [km]
## 6 134.3202 [km] 155.2134 [km] 223.9622 [km]
## DirectDistanceToOdense DirectDistanceToRoskilde DirectDistanceToBlegdamsvej
## 1 195.7070 [km] 246.0576 [km] 267.1295 [km]
## 2 194.1752 [km] 243.9046 [km] 264.9078 [km]
## 3 191.5805 [km] 242.8237 [km] 264.1505 [km]
## 4 194.2575 [km] 247.5975 [km] 269.2164 [km]
## 5 194.6926 [km] 249.4293 [km] 271.2758 [km]
## 6 190.0200 [km] 243.8173 [km] 265.6139 [km]
## DirectDistanceToGlostrup DirectDistanceToNearest Postcode
## 1 259.1978 [km] 64.51209 [km] 28310
## 2 256.9993 [km] 64.58528 [km] 28311
## 3 256.1223 [km] 60.81974 [km] 28312
## 4 261.0852 [km] 59.18484 [km] 28313
## 5 263.0624 [km] 57.41624 [km] 28314
## 6 257.4142 [km] 56.00828 [km] 2831511.4 Spatial regression
This is data published in Jama 29/4/2020 on COVD-19 in New York. The New York borough shapefiles were obtained from New York Open Data at https://data.cityofnewyork.us/City-Government/Borough-Boundaries/tqmj-j8zm. For those wishing to evaluate other datasets, there’s lung and lip cancer data in SpatialEpi library, leukemia in DClusterm library. Key aspect of spatial regression is that neighbouring regions are similar and distant regions are less so. It uses the polyn2nb in spdep library to create the neighbourhood weight. The map of residual for the New York data does not suggest spatial association of residuals.
The Moran’s I is used to test for global spatial autocorrelation among adjacent regions in multidimensional space. Moran’s I requires calculation of neighbourhood. It is related to the number of regions and sum of all spatial weights.
11.4.1 New York COVID-19 mortality
The example below illustrate the need to look at the data. It is not possible to perform spatial regression given the small size of the areal data (4 connected boroughs); Staten Island does not have adjoining border with the others nor share subway system. An intriguing possibility is that areal data analysis at the neighbourhood level would allow a granular examination of socioeconomic effect of COVID-19 on mortality data. To plot the railway lines with tmap the argument tm_lines is required whereas the argument tm_polygons is better suited for plotting the shape file. It
## Loading required package: spData## To access larger datasets in this package, install the spDataLarge
## package with: `install.packages('spDataLarge',
## repos='https://nowosad.github.io/drat/', type='source')`## Loading required package: Matrix##
## Attaching package: 'Matrix'## The following objects are masked from 'package:tidyr':
##
## expand, pack, unpack## Registered S3 methods overwritten by 'spatialreg':
## method from
## residuals.stsls spdep
## deviance.stsls spdep
## coef.stsls spdep
## print.stsls spdep
## summary.stsls spdep
## print.summary.stsls spdep
## residuals.gmsar spdep
## deviance.gmsar spdep
## coef.gmsar spdep
## fitted.gmsar spdep
## print.gmsar spdep
## summary.gmsar spdep
## print.summary.gmsar spdep
## print.lagmess spdep
## summary.lagmess spdep
## print.summary.lagmess spdep
## residuals.lagmess spdep
## deviance.lagmess spdep
## coef.lagmess spdep
## fitted.lagmess spdep
## logLik.lagmess spdep
## fitted.SFResult spdep
## print.SFResult spdep
## fitted.ME_res spdep
## print.ME_res spdep
## print.lagImpact spdep
## plot.lagImpact spdep
## summary.lagImpact spdep
## HPDinterval.lagImpact spdep
## print.summary.lagImpact spdep
## print.sarlm spdep
## summary.sarlm spdep
## residuals.sarlm spdep
## deviance.sarlm spdep
## coef.sarlm spdep
## vcov.sarlm spdep
## fitted.sarlm spdep
## logLik.sarlm spdep
## anova.sarlm spdep
## predict.sarlm spdep
## print.summary.sarlm spdep
## print.sarlm.pred spdep
## as.data.frame.sarlm.pred spdep
## residuals.spautolm spdep
## deviance.spautolm spdep
## coef.spautolm spdep
## fitted.spautolm spdep
## print.spautolm spdep
## summary.spautolm spdep
## logLik.spautolm spdep
## print.summary.spautolm spdep
## print.WXImpact spdep
## summary.WXImpact spdep
## print.summary.WXImpact spdep
## predict.SLX spdep##
## Attaching package: 'spatialreg'## The following objects are masked from 'package:spdep':
##
## anova.sarlm, as.spam.listw, as_dgRMatrix_listw, as_dsCMatrix_I,
## as_dsCMatrix_IrW, as_dsTMatrix_listw, bptest.sarlm, can.be.simmed,
## cheb_setup, coef.gmsar, coef.sarlm, coef.spautolm, coef.stsls,
## create_WX, deviance.gmsar, deviance.sarlm, deviance.spautolm,
## deviance.stsls, do_ldet, eigen_pre_setup, eigen_setup, eigenw,
## errorsarlm, fitted.gmsar, fitted.ME_res, fitted.sarlm,
## fitted.SFResult, fitted.spautolm, get.ClusterOption,
## get.coresOption, get.mcOption, get.VerboseOption,
## get.ZeroPolicyOption, GMargminImage, GMerrorsar, griffith_sone,
## gstsls, Hausman.test, HPDinterval.lagImpact, impacts, intImpacts,
## Jacobian_W, jacobianSetup, l_max, lagmess, lagsarlm, lextrB,
## lextrS, lextrW, lmSLX, logLik.sarlm, logLik.spautolm, LR.sarlm,
## LR1.sarlm, LR1.spautolm, LU_prepermutate_setup, LU_setup,
## Matrix_J_setup, Matrix_setup, mcdet_setup, MCMCsamp, ME, mom_calc,
## mom_calc_int2, moments_setup, powerWeights, predict.sarlm,
## predict.SLX, print.gmsar, print.ME_res, print.sarlm,
## print.sarlm.pred, print.SFResult, print.spautolm, print.stsls,
## print.summary.gmsar, print.summary.sarlm, print.summary.spautolm,
## print.summary.stsls, residuals.gmsar, residuals.sarlm,
## residuals.spautolm, residuals.stsls, sacsarlm, SE_classic_setup,
## SE_interp_setup, SE_whichMin_setup, set.ClusterOption,
## set.coresOption, set.mcOption, set.VerboseOption,
## set.ZeroPolicyOption, similar.listw, spam_setup, spam_update_setup,
## SpatialFiltering, spautolm, spBreg_err, spBreg_lag, spBreg_sac,
## stsls, subgraph_eigenw, summary.gmsar, summary.sarlm,
## summary.spautolm, summary.stsls, trW, vcov.sarlm, Wald1.sarlm##
## Attaching package: 'MASS'## The following objects are masked from 'package:raster':
##
## area, select## The following object is masked from 'package:dplyr':
##
## selectdfj<-data.frame(
Borough=c("Bronx","Brooklyn","Manhattan","Queens","Staten Island"),
Pop=c(1432132,2582830,1628701,2278906,476179),
Age65=c(12.8,13.9,16.5,15.7,16.2),
White=c(25.1,46.6,59.2,39.6,75.1),
Hispanic=c(56.4,19.1,25.9,28.1,18.7),
Afro.American=c(38.3,33.5,16.9,19.9,11.5),
Asian=c(4.6,13.4,14,27.5,11),
Others=c(36.8,10.4,15.4,17,5.2),
Income=c(38467,61220,85066,69320,82166),
Beds=c(336,214,534,144,234),
COVIDtest=c(4599,2970,2844,3800,5603),
COVIDhosp=c(634,400,331,560,370),
COVIDdeath=c(224,181,122,200,143),
COVIDdeathlab=c(173,132,91,154,117),
Diabetes=c(16,27,15,22,25),
Obesity=c(32,12,8,11,8),
Hypertension=c(36,29,23,28,25)) %>%
#reverse prevalence per 100000 to raw
mutate(Age65raw=round(Age65/100*Pop,0),
Bedsraw=round(Beds/100000*Pop,0),
COVIDtestraw=round(COVIDtest/100000*Pop,0),
COVIDhospraw=round(COVIDhosp/100000*Pop,0),
COVIDdeathraw=round(COVIDdeath/100000*Pop),0)
#Expected
rate<-sum(dfj$COVIDdeathraw)/sum(dfj$Pop)
dfj$Expected<-with(dfj, Pop*rate )
#SMR standardised mortality ratio
dfj$SMR<-with(dfj, COVIDdeathraw/Expected)
#NY Shape file - see this file open from above chunk
NYsf<-st_read("./Data-Use/Borough_Boundaries/geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0.shp")## Reading layer `geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\Borough_Boundaries\geo_export_7d3b2726-20d8-4aa4-a41f-24ba74eb6bc0.shp' using driver `ESRI Shapefile'
## Simple feature collection with 5 features and 4 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -74.25559 ymin: 40.49612 xmax: -73.70001 ymax: 40.91553
## geographic CRS: WGS84(DD)#join dataset
NYsf<-left_join(NYsf, dfj,by=c("boro_name"="Borough"))
#contiguity based neighbourhood
NY.nb<-poly2nb(NYsf)
is.symmetric.nb(NY.nb) # TRUE## [1] TRUE#NY subway
NYsubline<-st_read("./Data-Use/NYsubways/geo_export_147781bc-e472-4c12-8cd2-5f9859f90706.shp")## Reading layer `geo_export_147781bc-e472-4c12-8cd2-5f9859f90706' from data source `C:\Users\phant\Documents\Book\HealthcareRbook\Data-Use\NYsubways\geo_export_147781bc-e472-4c12-8cd2-5f9859f90706.shp' using driver `ESRI Shapefile'
## Simple feature collection with 742 features and 6 fields
## geometry type: LINESTRING
## dimension: XY
## bbox: xmin: -74.03088 ymin: 40.57559 xmax: -73.75541 ymax: 40.90312
## geographic CRS: WGS84(DD)#raw data
tm_shape(NYsf) +
tm_polygons(col='SMR',title='COVID raw') +
tm_shape(NYsubline)+tm_lines(col='name')The standardized mortality ratio (ratio of mortality divided by expected value for each borough) for the boroughs were: Bronx (1.245), Brooklyn (1.006), Manhattan (0.678) Queens (1.111) and Staten Island (0.794). The Figure shows a strong relationship between standardized mortality ratio and Income (R2=0.816).
#plot regression lines linear vs robust linear
ggplot(data=NYsf,aes(x=Income,y=COVIDdeath)) + geom_point() + geom_smooth(method='lm',col='darkblue',fill='blue') + geom_smooth(method='rlm',col='darkred',fill='red')## `geom_smooth()` using formula 'y ~ x'
## `geom_smooth()` using formula 'y ~ x'
Standardized mortality ratio and Income among different ethnic groups in Boroughs of New York.
#varies by ethinicty
dfj_long<-pivot_longer(data=dfj, names_to = "Ethnicity", values_to = "Popsize",cols=c(White,Afro.American,Asian,Hispanic,Others))
commonplot<-list(
scale_size_continuous(name = "Population size"),xlab("Income (Thousand)"),facet_wrap(~Ethnicity,nrow=2)
)
ggplot(data=dfj_long, mapping=aes(x=Income/1000, y=SMR, size=Popsize,colour=Borough))+geom_point()+commonplot
Robust regression to obtain residual for plotting with thematic maps.
#robust linear models
NYsf$resids <- rlm(COVIDdeathraw~Pop+Age65raw,data=NYsf)$res
#tmap robust linear model-residual
#plot using color blind argument
par(mfrow=c(2,1))
tm_shape(NYsf) + tm_polygons(col='resids',title='Residuals')## Variable(s) "resids" contains positive and negative values, so midpoint is set to 0. Set midpoint = NA to show the full spectrum of the color palette.## Variable(s) "resids" contains positive and negative values, so midpoint is set to 0. Set midpoint = NA to show the full spectrum of the color palette.#create spatial weights for neighbour lists
r.id<-attr(NYsf,"region.id")
lw <- nb2listw(NY.nb,zero.policy = TRUE) #W=row standardisedIn this example, the Moran’s I test suggest that the null test can be rejected. This implies random distribution of stroke across the regions of Denmark.
#globaltest spatial autocorrelation using Moran I test from spdep
gm<-moran.test(NYsf$SMR,listw = lw , na.action = na.omit, zero.policy = T)
gm##
## Moran I test under randomisation
##
## data: NYsf$SMR
## weights: lw n reduced by no-neighbour observations
##
##
## Moran I statistic standard deviate = 0.12086, p-value = 0.4519
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## -0.31010080 -0.33333333 0.03694814There’s no evidence of spatial autocorrelation at local level. Note that this data is too small and may not be the ideal to evaluate local Moran’s.
#local test of autocorrelation
lm<-localmoran(NYsf$SMR,listw = nb2listw(NY.nb, zero.policy = TRUE,
style = "C") , na.action = na.omit, zero.policy = T)
lm## Ii E.Ii Var.Ii Z.Ii Pr(z > 0)
## 1 -0.37737978 -0.2 0.1846888 -0.4127469 0.6601040
## 2 0.00000000 0.0 0.0000000 NaN NaN
## 3 -0.05282102 -0.2 0.1846888 0.3424724 0.3659977
## 4 0.03765408 -0.3 0.1729669 0.8118776 0.2084309
## 5 -1.25295769 -0.3 0.1729669 -2.2913539 0.9890285
## attr(,"call")
## localmoran(x = NYsf$SMR, listw = nb2listw(NY.nb, zero.policy = TRUE,
## style = "C"), zero.policy = T, na.action = na.omit)
## attr(,"class")
## [1] "localmoran" "matrix" "array"11.4.2 Danish Stroke Registry
We now return to spatial regression with the data from Danish stroke registry described above. First we will calculate the SMR
#Expected
rate<-sum(DKnuts2_sf2$strokenum)/sum(DKnuts2_sf2$pop)
DKnuts2_sf2$Expected<-with(DKnuts2_sf2, pop*rate )
#SMR standardised mortality ratio
DKnuts2_sf2$SMR<-with(DKnuts2_sf2, strokenum/Expected)
tm_shape(DKnuts2_sf2) +
tm_polygons(col='SMR',title='Stroke') Now we plot the neighborhood weight to see if the Zealand island is linked to Jutland peninsula. The data is in the .nb file. It looks like we may need to recalculate the neighborhood as there are bridges between Zealand and Zealand. This is different situation from Staten Island and the other boroughs of New York.
## Neighbour list object:
## Number of regions: 5
## Number of nonzero links: 6
## Percentage nonzero weights: 24
## Average number of links: 1.2## [1] TRUEPlotting in base R with sf object requires extracting the geometry and coordinates. Need to modify the link between Zealand and Jutland.
plot(st_geometry(DKnuts2_sf2))
plot(DKnut2.nb,coords=st_coordinates(st_centroid(st_geometry(DKnuts2_sf2))),
add=TRUE,pch=16,col='darkred')
The solution is provided in stack overflow. https://gis.stackexchange.com/questions/413159/how-to-assign-a-neighbour-status-to-unlinked-polygons
DKconnect <- function(polys, nb, distance="centroid"){
if(distance == "centroid"){
coords = sf::st_coordinates(sf::st_centroid(sf::st_geometry(polys)))
dmat = as.matrix(dist(coords))
}else if(distance == "polygon"){
dmat = sf::st_distance(polys) + 1 # offset for adjacencies
diag(dmat) = 0 # no self-intersections
}else{
stop("Unknown distance method")
}
gfull = igraph::graph.adjacency(dmat, weighted=TRUE, mode="undirected")
gmst = igraph::mst(gfull)
edgemat = as.matrix(igraph::as_adj(gmst))
edgelistw = spdep::mat2listw(edgemat)
edgenb = edgelistw$neighbour
attr(edgenb,"region.id") = attr(nb, "region.id")
allnb = spdep::union.nb(nb, edgenb)
allnb
}
#run function
DKnut2_connected.nb = DKconnect(DKnuts2_sf2,DKnut2.nb)
plot(st_geometry(DKnuts2_sf2))
plot(DKnut2_connected.nb,
coords=st_coordinates(st_centroid(st_geometry(DKnuts2_sf2))),
add=TRUE,pch=16,col='darkred')
#create spatial weights for neighbour lists
r.id<-attr(DKnuts2_sf2,"id")
lw <- nb2listw(DKnut2_connected.nb,zero.policy = TRUE) #W=row standardisedPerform robust regression
#robust linear models
DKnuts2_sf2$resids <- MASS::rlm(SMR~maleper,data=DKnuts2_sf2)$res
#tmap robust linear model-residual
#plot using color blind argument
tm_shape(DKnuts2_sf2) + tm_polygons(col='resids',title='Residuals')+
tm_style("col_blind")## Variable(s) "resids" contains positive and negative values, so midpoint is set to 0. Set midpoint = NA to show the full spectrum of the color palette.Check for spatial autocorrelation
#globaltest spatial autocorrelation using Moran I test from spdep
gm<-moran.test(DKnuts2_sf2$SMR,listw = lw ,
na.action = na.omit, zero.policy = T)
gm##
## Moran I test under randomisation
##
## data: DKnuts2_sf2$SMR
## weights: lw
##
## Moran I statistic standard deviate = -0.98413, p-value = 0.8375
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## -0.7354200 -0.2500000 0.2432931Local test of autocorrelation
#local test of autocorrelation
lm<-localmoran(DKnuts2_sf2$SMR,
listw = nb2listw(DKnut2_connected.nb, zero.policy = TRUE,
style = "C") , na.action = na.omit, zero.policy = T)
lm## Ii E.Ii Var.Ii Z.Ii Pr(z > 0)
## 1 -0.4777173 -0.15625 0.3396706 -0.55157917 0.7093816
## 2 -0.9220960 -0.15625 0.3396706 -1.31405173 0.9055856
## 3 -1.3492564 -0.31250 0.4705877 -1.51132003 0.9346465
## 4 -0.2490245 -0.31250 0.4705877 0.09253066 0.4631382
## 5 -0.1984676 -0.31250 0.4705877 0.16622945 0.4339882
## attr(,"call")
## localmoran(x = DKnuts2_sf2$SMR, listw = nb2listw(DKnut2_connected.nb,
## zero.policy = TRUE, style = "C"), zero.policy = T, na.action = na.omit)
## attr(,"class")
## [1] "localmoran" "matrix" "array"Spatial regression with spdep. The spatial filtering removes spatial dependency for regression analysis.
##spdep & spatialreg
fit.ols<-lm(SMR~maleper, data=DKnuts2_sf2,
listw=lw,zero.policy=T, type="lag", method="spam")
summary(fit.ols)SAR - Lag model
fit.lag<-lagsarlm(SMR~maleper, data=DKnuts2_sf2,
listw=lw,zero.policy=T, type="lag", method="spam")
summary(fit.lag, Nagelkerke=T)Spatial Durbin Model
#spatialreg
fit.durb<-lagsarlm(SMR~maleper,data=DKnuts2_sf2,
listw=lw,zero.policy=T, type="mixed", method="spam")
summary(fit.durb, Nagelkerke=T)Spatial Durbin Error Model
fit.errdurb<-errorsarlm(SMR~maleper, data=DKnuts2_sf2, listw=lw,zero.policy=T,etype="emixed", method="spam")
summary(fit.errdurb, Nagelkerke=T)SAC Model
fit.sac<-sacsarlm(SMR~maleper,data=DKnuts2_sf2,
listw=lw,zero.policy=T, type="sac", method="MC")
summary(fit.sac, Nagelkerke=T)spatial filtering
#function from spatialreg
#Set ExactEV=TRUE to use exact expectations and variances rather than the expectation and variance of Moran's I from the previous iteration, default FALSE
DKFilt<-SpatialFiltering(SMR~maleper,
data=DKnuts2_sf2,
nb=DKnut2_connected.nb,
#ExactEV = TRUE,
zero.policy = TRUE,style="W")11.4.3 INLA
This section uses Bayesian modeling for regression with fitting of the model by Integrated Nested Lapace Approximation (INLA). https://www.r-bloggers.com/spatial-data-analysis-with-inla/. For those wanting to analyse leukemia in New York instead of COVID-19, the dataset NY8 is available from DClusterm. INLA approximates the posterior distribution as latent Gaussian Markov random field. In this baseline analysis, the poisson model is performed without any random effect terms.
library(INLA)
nb2INLA("DKnut2.graph", DKnut2_connected.nb)
#This create a file called ``LDN-INLA.adj'' with the graph for INLA
DK.adj <- paste(getwd(),"/DK.graph",sep="")
#Poisson model with no random latent effect-ideal baseline model
m1<-inla(SMR~ 1+maleper, data=DKnuts2_sf2, family="poisson",
E=DKnuts2_sf2$Expected,control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE), verbose = T )
R1<-summary(m1)In this next analysis, the Poisson model was repeated with random effect terms. This step was facilitated by adding the index term.
#Poisson model with random effect
#index to identify random effect ID
DKnuts2_sf2$ID <- 1:nrow(DKnuts2_sf2)
m2<-inla(SMR~ 1+ maleper +f(ID, model = "iid"), data=DKnuts2_sf2, family="poisson",
E=DKnuts2_sf2$Expected,control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE) )
R2<-summary(m2)
DKnut2_sf2$FIXED.EFF <- m1$summary.fitted[, "mean"]
DKnut2_sf2$IID.EFF <- m2$summary.fitted[, "mean"]
#plot regression on map
tSMR<-tm_shape(DKnuts2_sf2)+tm_polygons("SMR")
tFIXED<-tm_shape(DKnuts2_sf2)+tm_polygons("FIXED.EFF")
tIID<-tm_shape(DKnuts2_sf2)+tm_polygons("IID.EFF")This next paragraph involves the use of spatial random effects in regression models. Examples include conditional autoregressive (CAR) and intrinsic CAR (ICAR).
# Create sparse adjacency matrix
DK.mat <- as(nb2mat(DKnut2_connected.nb,
style = "B",zero.policy = TRUE),"Matrix")
#S=variance stabilise
# Fit model
m.icar <- inla(SMR ~ 1+maleper+
f(ID, model = "besag", graph = DK.mat),
data = DKnuts2_sf2, E = DKnuts2_sf2$Expected, family ="poisson",
control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE))
R3<-summary(m.icar)The Besag-York-Mollie (BYM) now accounts for spatial dependency of neighbours. It includes random effect from ICA and index.
m.bym = inla(SMR ~ -1+ maleper+
f(ID, model = "bym", graph = DK.mat),
data = DKnuts2_sf2, E = DKnuts2_sf2$Expected, family ="poisson",
control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE))
R4<-summary(m.bym)ICARmatrix <- Diagonal(nrow(DK.mat), apply(DK.mat, 1, sum)) - DK.mat
Cmatrix <- Diagonal(nrow(DKnuts2_sf2), 1) - ICARmatrix
max(eigen(Cmatrix)$values)
m.ler = inla(SMR ~ -1+maleper+
f(ID, model = "generic1", Cmatrix = Cmatrix),
data = DKnuts2_sf2, E = DKnuts2_sf2$Expected, family ="poisson",
control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE))
R5<-summary(m.ler)Spatial econometric model usch as spatial lag model includes covariates and autoregres on the response variable.
#X
mmatrix <- model.matrix(SMR ~ 1, DKnuts2_sf2)
#W
W <- as(nb2mat(DKnut2_connected.nb, style = "W", zero.policy = TRUE), "Matrix")
#Q
Q.beta = Diagonal(n = ncol(mmatrix), x = 0.001)
#Range of rho
rho.min<- -1
rho.max<- 1
#Arguments for 'slm'
args.slm = list(
rho.min = rho.min ,
rho.max = rho.max,
W = W,
X = mmatrix,
Q.beta = Q.beta
)
#Prior on rho
hyper.slm = list(
prec = list(
prior = "loggamma", param = c(0.01, 0.01)),
rho = list(initial=0, prior = "logitbeta", param = c(1,1))
)
#SLM model
m.slm <- inla( SMR ~ -1+maleper+
f(ID, model = "slm", args.slm = args.slm, hyper = hyper.slm),
data = DKnuts2_sf2, family = "poisson",
E = DKnuts2_sf2$Expected,
control.predictor = list(compute = TRUE),
control.compute = list(dic = TRUE, waic = TRUE)
)
R6<-summary(m.slm)
marg.rho.internal <- m.slm$marginals.hyperpar[["Rho for ID"]]
marg.rho <- inla.tmarginal( function(x) {
rho.min + x * (rho.max - rho.min)
}, marg.rho.internal)
inla.zmarginal(marg.rho, FALSE)
plot(marg.rho, type = "l", main = "Spatial autocorrelation")Spatial model selection
DKnut2_sf2$ICAR <- m.icar$summary.fitted.values[, "mean"]
DKnut2_sf2$BYM <- m.bym$summary.fitted.values[, "mean"]
DKnut2_sf2$LEROUX <- m.ler$summary.fitted.values[, "mean"]
DKnut2_sf2$SLM <- m.slm$summary.fitted.values[, "mean"]
labels<-c("Fixed","IID", "ICAR","BYM","LEROUX","SLM")
Marginal_Likelihood<-c(R1$mlik[1],R2$mlik[1],R3$mlik[1],R4$mlik[1],R5$mlik[1],
R6$mlik[1])
Marginal_Likelihood<-round(Marginal_Likelihood,2)
WAIC<-c(R1$waic[[1]],R2$waic[[1]],R3$waic[[1]],R4$waic[[1]],R5$waic[[1]],
R6$waic[[1]])
WAIC<-round(WAIC,2)
DIC<-c(R1$dic[[1]],R2$dic[[1]],R3$dic[[1]],R4$dic[[1]],R5$dic[[1]],R6$dic[[1]])
DIC<-round(DIC,2)
Results<-data.frame(labels,Marginal_Likelihood,WAIC,DIC)
knitr::kable(Results)
#plot maps
tICAR<-tm_shape(DKnut2_sf2)+tm_polygons("ICAR")
tBYM<-tm_shape(DKnut2_sf2)+tm_polygons("BYM")
tLEROUX<-tm_shape(DKnut2_sf2)+tm_polygons("LEROUX")
tSLM<-tm_shape(DKnut2_sf2)+tm_polygons("SLM")
#arrange in grid using tmap arrange
current.mode <- tmap_mode("plot")
tmap_arrange(tFIXED,tIID,tICAR,tBYM,tLEROUX,tSLM)
tmap_mode(current.mode)11.4.4 Stan
## Loading required package: Rcpp## Warning: package 'Rcpp' was built under R version 4.0.5## Loading 'brms' package (version 2.14.4). Useful instructions
## can be found by typing help('brms'). A more detailed introduction
## to the package is available through vignette('brms_overview').##
## Attaching package: 'brms'## The following object is masked from 'package:dismo':
##
## kfold## The following object is masked from 'package:stats':
##
## ar11.5 Machine learning in spatial analysis
Up until now we have used frequentist and Bayesian spatial regression methods. Spatial data can also be analysed using machine learning such as random forest.
11.6 Spatio-temporal regression
Spatio-temporal regression combines a spatial model with a temporal model. In many cases of low disease incidene in each region it may not be possible to identify any temporal trend at a regional level.