# source.folder <- "/full/path/NeMo_output"
source.folder <- "/Users/au301842/NeMo_output"6 ZIP to data set
So, here I’ll show how we went from compressed zip file, from the NeMo tool, and to having a data set for further analyses.
I am fairly new to Python, but have worked in R for many years. This means, that I have written just enough in Python, and everything else in R. Small python functions will be wrapped in R scripts. Just a word of warning.
First, extract the compressed file and put it where you like. I’ll denote this location as the source.folder.
Then we want to extract the chacovol data. This is extracted by unpikling .pkl files using a short python script wrapped in R:
source("nemo/unpkl-sparse.R")
unpkl_sparse(data.folder = source.folder,
"chacovol_yeo17_mean.pkl")The unpikled files are then collected and merged in a wide format.
But first, we need to handle the provided atlas .txt files to get ROI names:
atlas <- readLines("/Users/au301842/NeMo_output/Yeo2011_17Networks_NetworkNames_ColorLUT.txt")
rois <- do.call(c,lapply(atlas[-1],function(i){
s1 <- strsplit(i,"_[0-9]{1,3}_")[[1]][2]
strsplit(s1," ")[[1]][1]
}
))source("nemo/nemo-collect.R")
df <- nemo_collect(
data.folder = source.folder,
id.pattern = "W[0-9]{2}",
file.pattern = "chacovol_yeo17_mean.tsv",
roi.names = rois
)I very much prefer writing function to do the data handling, but I just have to note, that these functions are very primitive due to lack of time on my end. But they are working and will provide a good foundation for further worker.
6.1 Visualisation
This means it is technically not allowed to modify or redistribute the code. Please refer to {#contents}.
We also want those nice glass brains. These following R scripts are all simple wrappers for the scripts from the NeMo tool, which uses the nibabel Python package for plotting. All the examples below are created based on a small sample of two subjects and the Yeo17 atlas which is also shared in the NeMo tool source.
First we do the simpel voxel based heatmap.
source("nemo/glass-brain.R")
glass_brain(
data.folder = source.folder,
file.pattern = "chacovol_res2mm_mean.nii.gz",
out.name = "images/glass_chacovol.png"
)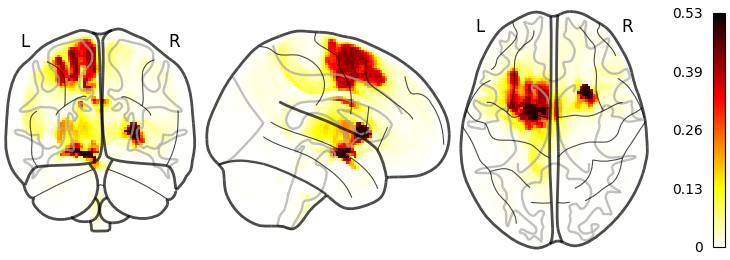 Next we can do the parcellation or atlas based chacovol plotting:
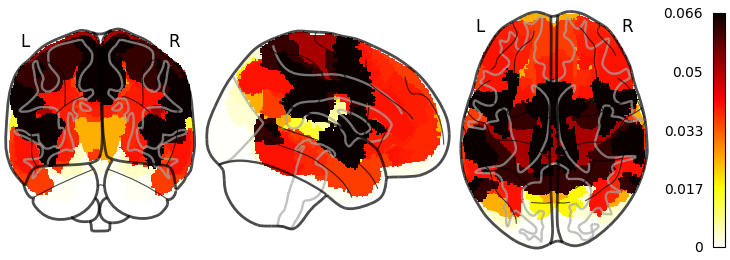
Next we can do the parcellation or atlas based chacovol plotting:
source("nemo/glass-brain.R")
glass_brain(
data.folder = source.folder,
file.pattern = "chacovol_yeo17_mean.pkl",
out.name = "images/glass_chacovol_parc.png",
parcellation = "/Users/atlas/folder/Yeo2011_17Networks_MNI152_182x218x182_LiberalMask.nii.gz"
)
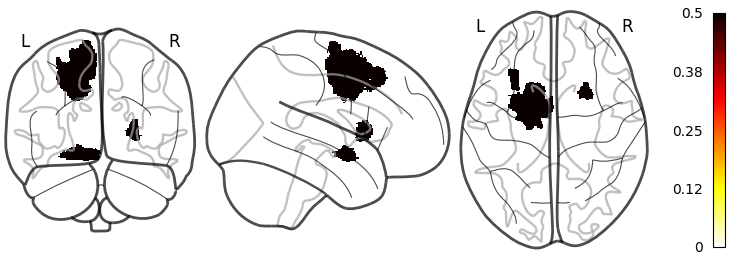
The same scirpt can also be used for lesion plotting:
glass_brain(
data.folder = "/Users/folder/with/lesions/",
file.pattern = "lesion.nii.gz",
id.pattern = "W(50|49)", # This id.pattern is used to only include specified ids
out.name = "images/glass_lesion.png"
)
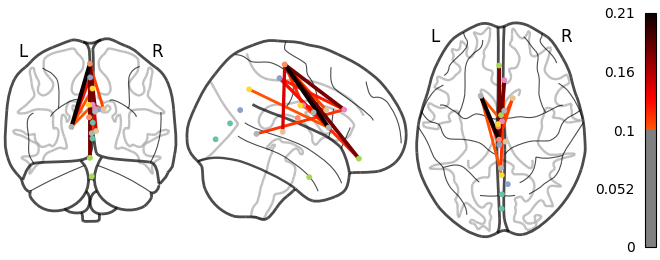
And finally the change in connections between areas in the brain can be visualised as a graphbrain:
source("nemo/graph-brain.R")
graph_brain(
data.folder = source.folder,
file.pattern = "chacoconn_yeo17_mean.pkl",
out.name = "images/glass_chacoconn.png",
node.file="/Users/atlas/folder/Yeo2011_17Networks_MNI152_182x218x182_LiberalMask.nii.gz"
)
6.2 Glossary
data frame with 0 columns and 0 rows